
Guidance on whole genome sequencing in the analysis of microorganisms
In 2021, the EFSA (European Food Safety Authority) released a statement concerning the use of whole genome sequencing (WGS) in the examination of microorganisms deliberately utilized in the food chain. This statement offers guidelines for conducting WGS analysis and evaluating risks tied to microorganisms in food production and processing. The aim is also to bring about standardized practices in this sector.
The QPS concept
In aiding the risk assessment of microorganisms, EFSA introduced the qualified presumption of safety (QPS) concept. for microorganisms as a generic pre-assessment approach that covers safety concerns for humans, animals, and the environment. This acts as a generic pre-assessment method covering safety concerns for humans, animals, and the environment. Within the QPS framework, a safety review for a specific taxonomic unit (TU) is performed independently during market authorisation. Strains belonging to QPS TUs still require an assessment based on a specific data package reviewed by the relevant EFSA Panel. Ideally, QPS status should expedite this evaluation.
Safety concerns for a TU are categorised as ‘qualifications’, assessed at the strain level. A general qualification is that "strains should not harbour any acquired antimicrobial resistance genes to clinically relevant antimicrobials". This applies to all QPS bacterial TUs. However, definitions of ‘acquired’ and ‘intrinsic’ AMR genes have caused some ambiguity in the interpretation.
The safety qualifications for a TU are categorized as 'qualifications', assessed at the strain level. A general qualification is that strains shouldn't possess any acquired antimicrobial resistance genes that are clinically relevant. However, definitions of ‘acquired’ and ‘intrinsic’ AMR genes have caused some ambiguity.
Proposal for interpretation of acquired and intrinsic AMR
In October 2023, EFSA released a statement on how to interpret the QPS qualification on ‘Acquired antimicrobial resistance genes. This statement proposed a bioinformatics strategy to determine the inherent or acquired nature of an AMR gene. All AMR genes conferring resistance towards important antimicrobials (IAs, CIAs or HIAs as defined by WHO 2019), found as hits in the bioinformatic analysis, need to be further evaluated. Genes conferring ‘intrinsic’ resistance could be considered as of no concern in the frame of the EFSA risk assessment. Acquired AMR genes coinciding with the corresponding antimicrobial phenotypic resistance should be considered as a concern. If the presence of the ‘acquired’ AMR gene is not leading to phenotypic resistance, a case-by-case assessment is necessary (Figure 1).
| Both approaches are combined |
MIC > cut-off | MIC ≤ cut-off |
| Gene found | HAZARD | Further studies to determine whether the gene may become active |
| Gene not found | Uncertainty: case-by-case assessment | OK |
Figure 1. Interpretation of the phenotypic and genotypic findings in antimicrobial resistance.
The use of a QPS microorganism, or its products, should not add to the overall pool of AMR genes or increase the spread of AMR. One primary consideration is the potential transferability of an AMR gene. The analysis of mobile genetic elements in the WGS of a bacterium and experiments for testing the transferability of genes are considered not applicable as a basis for the safety assessment, due to difficulties in reproducing the events in lab-designed experiments and potential false positive and false negative results due to DNA sequence bias and technical problems in bioinformatic analysis.
Instead, the statement focuses on the "intrinsic" or "acquired" nature of the hits corresponding to AMR genes identified from the WGS as explained in the EFSA (2021) statement. Here, the AMR genes are classified for the purpose of EFSA’s risk assessments, and they apply to bacteria used in the food and feed chains (Figure 2).
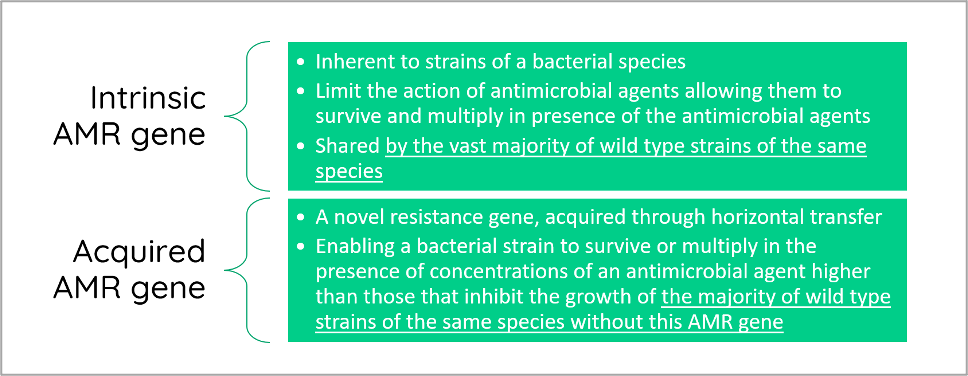
Figure 2. Definitions for intrinsic and acquired AMR genes according to EFSA.
The EFSA statement outlines the procedural steps and quality control measures essential for a bioinformatic risk assessment of AMR genes in bacterial strains using WGS data. Generally, the analysis should start with all sequences of the strains of the species available in the repositories. If fewer than 30 strains are available, it is recommended to include more strains based on the applicant's own research. For this research, high-quality draft genomes that meet the quality criteria set in the EFSA WGS statement (2021) are preferred. The species identity of the strains should also be verified, and the strains should also be independent isolates belonging to the same species. The non-clonal relationship needs to be demonstrated by a phylogenomic approach (Figure 3).
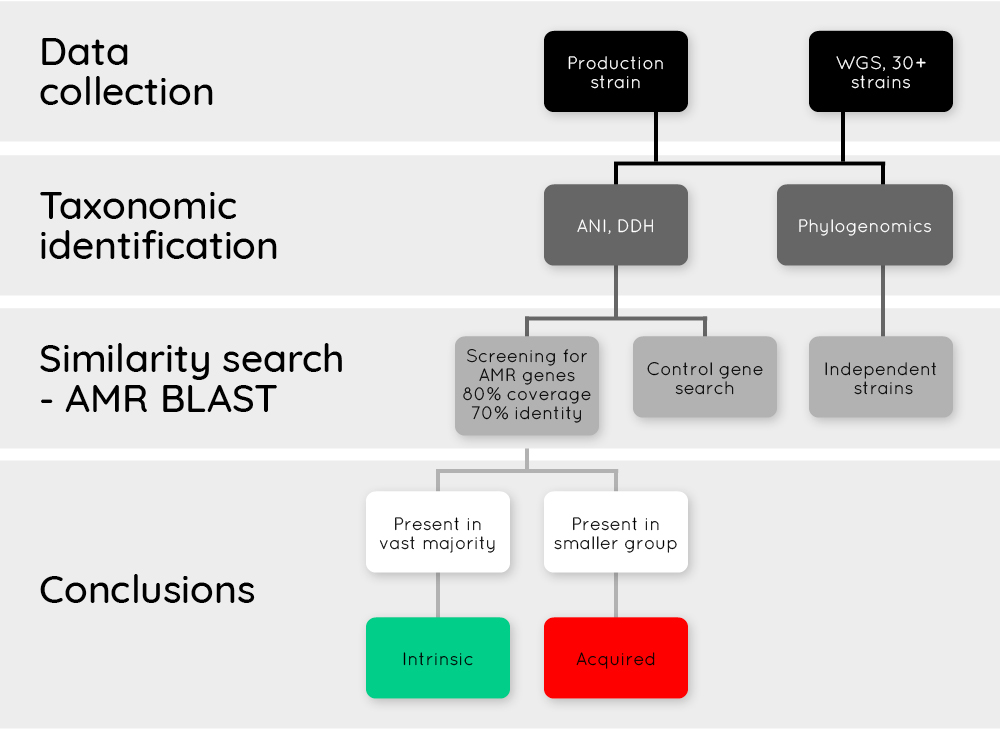
Figure 3. Flowchart of AMR interpretation.
The AMR assessment should be based on a similarity search (e.g. BLAST) of the collected WGS sequences for the presence of homologous AMR genes. For this assessment, the gene sequence as present in the production strain should be used. A positive control gene expected to be present in all strains of the target group is required. In general, query sequence hits with at least 80% identity (at the protein level or nucleotide level as reported in the database) and 70% length of the subject sequence should be considered. Hits identifying genes conferring "intrinsic" resistance could be considered as "of no concern". Acquired AMR genes with corresponding antimicrobial phenotypic resistance should be considered as a concern. If the presence of the "acquired" AMR gene is not leading to phenotypic resistance, a further case-by-case assessment is required.
Biosafe commented on the draft statement during the public commentary round, expressing gratitude for the unifying efforts on interpreting intrinsic and acquired resistance. Some concerns were raised, especially regarding the requirement to evaluate 30+ independent isolates, where it might pose a challenge to obtain WGS data from so many independent isolates. The laboratory strains are often derived from a single strain, and are thus phylogenomically considered as one, even if the species is well represented in public sequence databases. For rare species, the situation may be even worse, as the isolation and characterisation of a large number of strains is slow and costly, sometimes impossible.
Additionally, even though the EFSA statement does not recommend assessing the intrinsic or acquired nature of AMR genes based on the presence of mobile genetic elements, due to the lack of reliability in the bioinformatic tools, at least some of those could be considered as well. We would still consider for example including plasmid searches in the AMR risk assessment. Also, the "vast majority" concept could be defined to ensure the predictability of the risk assessment.
The statement was published on 31 October 2023, with an adoption date of 27 September 2023.
For more information:

Jenny Makkonen
Project Manager, PhD
+358 40 547 4110
jenny.makkonen@biosafe.fi
Author:




/Lopputuote/microbial-products-biosafe-2-1920x1280.jpg?width=540&name=microbial-products-biosafe-2-1920x1280.jpg)


/Lopputuote/microbial-products-cell-cultivated-meat-2-biosafe-1920x1280.jpg?width=540&name=microbial-products-cell-cultivated-meat-2-biosafe-1920x1280.jpg)

