Europe's leading CRO in
food and feed safety
Europe's leading Contract Research Organization in the safety assessment of microbial food and feed products.

Your end-to-end strategic CRO partner in biotech

"Navigating the complexities of EU food safety regulations can be overwhelming. The laboratory testing services provided by Biosafe are thorough and accurate, giving us peace of mind that our product, Solein, meets the highest standards."
— Juha-Pekka Pitkänen, CTO, Solar Foods

"CROs are well suited to the role of biotech 'thought partner', complementing biotech companies’ scientific expertise with their established relationships, their access to best-in-breed tools, and the sheer scale of their development engine."
— McKinsey, CROs and biotech companies: Fine-tuning the partnership
Biosafe helps new sustainable food & feed solutions get approved and reach the European market. From laboratory to legislation to the latest intelligence.

Genome sequencing and bioinformatics
Our professionals are vastly experienced in whole-genome sequencing and our pipeline is optimized to fulfil the safety assessment requirements of EFSA whether it's analyzing DNA for GMOs, AMR or anything else.

EU regulatory consulting
We have extensive expertise and experience in what the EU, the European Commission and EFSA demand of the safety of industrial microbiology products. Our consulting team can advise you on a wide range of topics on food safety and guide what you should be doing next.
Laboratory services

Find the latest news and insights from the world of food safety here.
Find your path to food safety — subscribe to our newsletter!
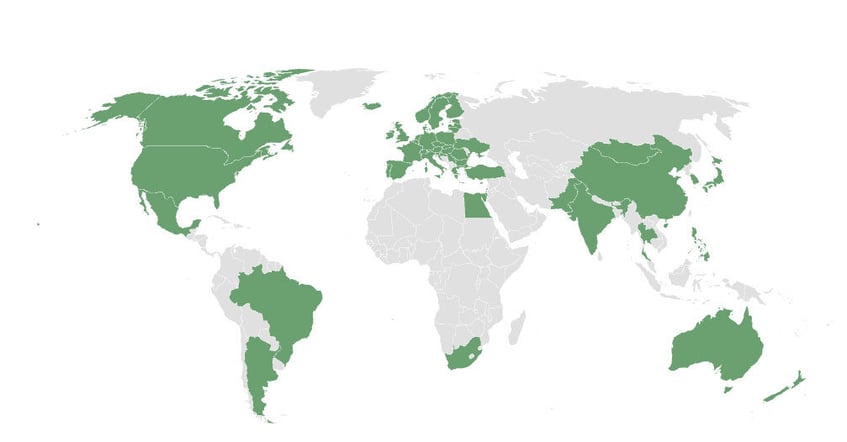
We help our customers develop safer products and solutions — all around the world.
See our all-inclusive guide to getting your product assessed for safety and approved for launch.
How people describe us
"Biosafe provides great extra ideas with professional answers, analysis methods, and research on literature when necessary. They always search for any required information and make sure it is accepted."
"I appreciate the ability to have an open discussion with the Biosafe team members. We share details, opinions, and the understanding to advance together on mutual terms."
"Biosafe has a great team of scientists who you can build a long-term relationship with."